

About Me!
Hi, I am Richa Mudgal, Ph.D and I am currently working as a Senior Manager - Bioinformatics at Elucidata, India. I am a self-motivated and diligent computational biologist with experience in multi-omics data analysis and building machine learning models for biological systems. My area of research can be broadly described as developing statistical and computational models for omics data for transforming drug discovery.
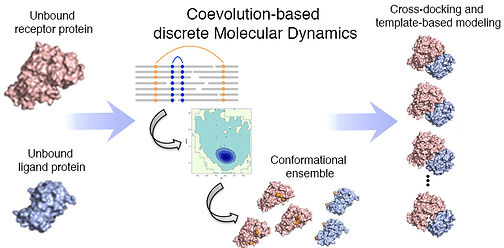
Before starting my stint in industry, I pursued postdoctoral research at the Institute for Research in Biomedicine (IRB), Barcelona, Spain, where I analyzed how evolutionary information can aid in sampling the transition from the unbound to the bound state, which further aids in the accurate modeling of protein-protein interactions.
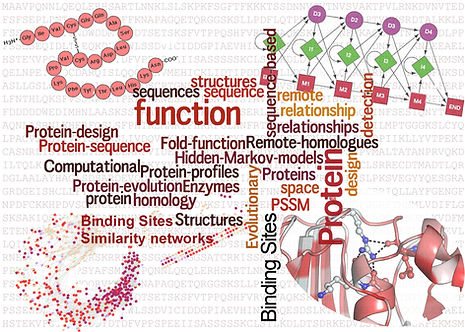
I completed my doctoral thesis in 2015 from Indian Institute of Science, Bangalore, India, which primarily focused on designing protein-like sequences and functional annotation of proteins with unknown functions. We also explored the relationship between enzyme folds and their function from a network perspective.
I am originally from New Delhi, India. In my free time, I enjoy travelling and watching TEDx talks and Kurzgesagt.
Work Experience
Sr. Manager Bioinformatics
ML & Insights Solutions
Elucidata, Delhi
Sept 2023 - Present
-
Managed 15+ customer projects specializing in omics and clinical data harmonization, single-cell analysis, gene similarity searches, building knowledge graphs, and predictive ML models.
-
Pioneered the development of clinical trials data curation, creating a new service offering that facilitated market expansion.
-
Implemented a data-driven QC metric that enhanced the public data selection process efficiency by 30%.
Solutions Manager
Data Harmonisation & Insights Solutions
Elucidata, Delhi
Jan 2022 - Sept 2023
-
Orchestrated technical leadership and imparted key insights, ensuring elevated customer satisfaction for over 13 customer accounts, leading to a revenue of $1.2M.
-
Managed a team of 10+ bioinformatics scientists and engineers.
-
Created 5+ templatized solutions and scalable processes to ensure timely and high-quality deliveries.
-
Facilitated strategic insights and forward-thinking approaches to expand existing accounts and minimize churn, ensuring a 100% Net Revenue Retention rate.
-
Collaborated closely with the product team, providing actionable feedback and insights based on customer needs analysis, resulting in a 20% increase in customer satisfaction scores.
-
Strategically hired 10+ members to bridge the technical gaps in delivery team.
Sr. Bioinformatics Scientist
Customer Success
Elucidata, Delhi
June 2019 - Dec 2021
-
Hands-on experience with processing and analyzing single-cell RNA-seq, Perturb-seq, CITE-seq, bulk transcriptomics, metabolomics and performing multi-omics, multi-modal and meta-analysis.
-
Designed a novel machine learning application (Polly- PeakML) for performing accurate peak detection and improving the untargeted metabolomics analysis workflows.
-
Led an innovations project for identifying drug combinations for infectious diseases specifically COVID-19, achieving the final phase of Drug Discovery Hackathon - 2020)
-
Authored scientific reports for FDA approval and developed a template for cataloging off-target effects for genes of interest for shRNAs.
Drug Discovery Hackaton 2020

Education

Thesis title: Inferences on structure and function of proteins from sequence data: Development of methods and applications.
Thesis supervisors: Prof N. Srinivasan and Prof. N Chandra.

M.Sc. Bioinformatics
Jamia Millia Islamia, India
2002-2005
80.8% (Gold Medalist)

B.Sc (Hons) Applied Zoology
Delhi University, India
2002-2005
77.62% (Ranked II in Delhi University)

Schooling
Modern School Vasant Vihar, New Delhi, India
Senior Secondary School Examination - 78%
Secondary School Examination - 87%
Research Interests

Biological Network Analysis
Personalised medicine

Drug repurposing and drug combinations

Protein Evolution

Protein sequence, structure and function relationships
Molecular mechanisms of Aging
and age-related diseases

Research Experience
Postdoctoral Research
Institute for Research in Biomedicine (IRB), Barcelona, Spain
Nov 2015 - Dec 2017
Postdoc supervisor - Prof. Patrick Aloy
Exploring the applicability of conformational ensembles generated using intrinsic coevolving residue pairs for modelling protein transitions upon complex formation.
PhD. in computational Biology
Indian Institute of Science, Bangalore, India
2008 - 2015
Thesis title: Inferences on structure and function of proteins from sequence data: Development of methods and applications.
Thesis supervisors: Prof N. Srinivasan and Prof. N Chandra.
Project Consultant
National Institute of Immunology, New Delhi, India
2007-2008
Worked with Dr. Sagar Sengupta in the project titled “(NIH GRIP) Regulatory Mechanisms for BLM helicase.”
Profile: - Provided Bioinformatics perspective to the projects, primarily prediction of phosphorylation sites in helicase proteins, structural approach to predict mitochondrial localization of helicase proteins and microarray data analysis to identify differentially expressed genes in BLM knockdown cells. Also had hands on experience on basic wet lab techniques.
(1) National Institute of Immunology, New Delhi, India
Supervisors: Dr. Sagar Sengupta and Dr. Debasisa Mohanty
Jan 2007 - July 2007 (Master's dissertation)
(2) Institute of Genomics and Integrative Biology, New Delhi, India
Supervisor: Dr. B. K. Malik
Jan 2006- July 2006 (Summer dissertation)
Selected Projects
Conformational sampling of proteins using intrinsic coevolutionary information and its application in improving modelling of protein-protein interactions

Publications
-
Kumar G., Mudgal R., Srinivasan N., and Sankaran S. (2018) Use of designed sequences in protein structure recognition. Biology Direct 13:8
-
Mudgal R., Srinivasan N. and Chandra N. (2017) Resolving protein structure-function-binding site relationships from a binding site similarity network perspective Proteins, doi:10.1002/prot.25293
-
Sandhya S., Mudgal R., Kumar G., Sowdhamini R. and Srinivasan N. (2016) Protein sequence design and its application. Current Opinion in Structural Biology 37, 71-80.
-
Mudgal R., Sandhya S., Chandra N. and Srinivasan N. (2015) De-DUFing the DUFs: Deciphering distant evolutionary relationships of DUFs using sensitive homology detection methods. Biology Direct 10:38.
-
Ramakrishnan G., Ochoa-Montaño B., Upadhyayula R.S., Mudgal R., Joshi A.G., Chandra N.R., Sowdhamini R., Blundell T.L. and Srinivasan N. (2014) Enriching the annotation of Mycobacterium tuberculosis H37Rv proteome using remote homology detection approaches: Insights into structure and function. Tuberculosis, Vol. 5 (1): 14-25
-
Mudgal R., Sandhya S., Kumar G., Sowdhamini R., Chandra N. and Srinivasan N. (2014) NrichD database: sequence databases enriched with computationally designed protein-like sequences aid in remote homology detection. Nucleic Acids Research, Vol. 43 (Database issue): D300-305.
-
Mudgal R., Sowdhamini R., Chandra N., Srinivasan N. and Sandhya S. (2014) Filling-in void and sparse regions in protein sequence space by protein-like artificial sequences enables remarkable enhancement in remote homology detection capability. Journal of Molecular Biology, Vol. 426(4): 962-979.
-
Chandra S., Priyadarshini R., Madhavan V., Tikoo S., Hussain M., Mudgal R., Modi P., Srivastava V. and Sengupta S. (2013) Enhancement of c-Myc degradation by Bloom (BLM) helicase leads to delayed tumor initiation. Journal of Cell Science, Vol. 126: 3782-3795.
-
Sandhya S., Mudgal R., Jayadev C., Abhinandan K.R., Sowdhamini R. and Srinivasan N. (2012) Cascaded walks in protein sequence space: use of artificial sequences in remote homology detection between natural proteins. Molecular BioSystems, Vol. 8: 2076–2084
-
Ramakrishnan G., Gowri V.S., Mudgal R., Chandra N.R. and Srinivasan N. (2012) Mining the sequence databases for homology detection: Application to recognition of functions of Trypanosoma brucei brucei proteins and drug targets. Biological Data Mining and Its Applications in Healthcare (Book Chapter).
-
De S., Kumari J., Mudgal R., Modi P., Gupta S., Futami K., Goto H., Lindor N.M., Furuichi Y., Mohanty D. and Sengupta S. (2012) RECQL4 is essential for the transport of p53 to mitochondria in normal human cells in the absence of exogenous stress. Journal of Cell Science, Vol. 125: 2509–2522.
-
Srinivasan N., Agarwal G., Bhaskara R.M., Gadkari R., Krishnadev O., Lakshmi B., Mahajan S., Mohanty S., Mudgal R., Rakshambikai R., Sandhya S., Sudha G., Swapna L.S. and Tyagi N. (2010) Influence of genomic and other biological data sets in the understanding of protein structures, functions and interactions. Int. J. Knowledge Discovery Bioinf. Vol. 2(1): 24-44.
-
Kaur S.*, Modi P.*, Srivastava V.*, Mudgal R.*, Tikoo S., Arora P., Mohanty D. and Sengupta S. (2010) Chk1-dependent constitutive phosphorylation of BLM helicase at Serine 646 decreases after DNA damage. Molecular Cancer Research, Vol. 8: 1234-1247 (* signifies equal first authors).
-
Srivastava S., Modi P., Tripathi V., Mudgal R., De S. and Sengupta S. (2009) BLM helicase stimulates the ATPase and chromatin remodeling activities of RAD54. Journal of Cell Science, Vol 122: 3093-3103.
-
Mudgal R., Sfriso P., Duran-Frigola M., Collisci F., Pallara C., Orozco M., Fernandex-Recio J., and Aloy P. (2017) Evolutionary driven conformational sampling improves detection of protein interactions (manuscript under preparation).
Database updates, online publications and conference abstracts
-
Mudgal R., Krishnadev O., Mukherjee S. and Srinivasan N. (2010, 2011, 2012, 2013) SUPFAM, http://www.oxfordjournals.org/nar/database/summary/219
-
Mohanty S., Mudgal R., Krishnadev O. and Srinivasan N. (2010) MulPSSM, http://www.oxfordjournals.org/nar/database/summary/844
-
Srinivasan N., Mudgal R., Kumar G., Chandra N.R., Sowdhamini R. and Sandhya S. (2015) 161. Bridging the islands of protein families in sequence space using artificial sequences. Journal of Biomolecular and Structural Dynamics, 33 Suppl 1:103-4. (Abstract book – Albany 2015
Other achievements
Participated in CASP 10 (2012) Competition under the category of template-based and ab-initio structure modeling and secured rank 51.
Group name: sumalab
Selected workshops and conferences
1. Participated and presented a poster titled "Improving search space for protein-protein docking using alternative functional conformations" in the MPI (Modeling of protein interactions) 2016 held from 27-29 October 2016, at Lawrence, Kansas, USA.
2. Participated and presented a poster titled "Tracing the similarities between enzyme folds through binding-site similarity networks" in the ISMB/ECCB 2013 conference Berlin (21st Annual Conference on Intelligent Systems for Molecular Biology (ISMB) and 12th European Conference on Computational Biology) held from 19-23 July 2013, at ICC, Berlin, Germany
3. Participated and presented a poster titled "Bridging the islands in protein sequence space through computationally designed protein-like sequences" in the "International Conference on Biomolecular Forms and Functions - 50 years of the Ramachandran Map" help from 8-11 January 2013, at Indian Institute of Science, Bangalore.
4. Participated in the "International School and Conference on "Networks in Biology, Social Science and Engineering" held from July 02 - 14, 2012 Indian Institute of Science, Bangalore.
5. Presented a poster titled “Walking in protein sequence space through directed design of protein-like sequences” at the International Conference on Mathematical Biology, July 04-07, 2011, Indian Institute of Science, Bangalore.
6. Participated in the workshop on "Introduction to Mathematical Techniques in Life Sciences" held during the period of January 1-4, 2011 at Indian Institute of Science, Bangalore.
7. Presented a poster titled “Directed computational design of protein-like sequences using probabilistic model of amino acid substitutions” at the The Eight Asia-Pacific Bioinformatics Conference, 2010
Online courses
-
Distinction in Stillwater’s Leadership Effectiveness Program
-
Coursera –
-
(1) Genomic and Precision Medicine by University of California, San Francisco (with Distinction)
-
(2) Learn to Program: The Fundamentals of Python, by University of Toronto (with Distinction)
-
(3) Computing for Data Analysis using R, by Johns Hopkins University (with Distinction)
-
(4) Introduction to Genetics and Evolution by Duke University (with Distinction)
-
(5) Network analysis in Systems Biology (Icahn School of Medicine at Mount Sinai - Audit)
-
-
Udemy –
-
(1) Python for Data Science and Machine Learning Bootcamp (certificate)
-
-
Linkedin –
-
(1) Data Science Foundations: Fundamentals (certificate)
-
Other Interests
 |  |
|---|---|
 |  |





